Abstract
Background
CD34, a transmembrane sialoglycoprotein, is expressed in hematopoietic stem and progenitor cells, endothelial cells, and bone marrow stromal cells. Therefore, in the clinical diagnosis and classification of leukemia, high expression of CD34 is customized as a marker for the immature phenotype. Although alternative splicing is a common oncogenic mechanism in various cancers, abnormal splicing of CD34 has not been revealed in hematological malignancies.
Methods
Here, we investigated the transcriptional profile of CD34, including expression level and alternative splicing, by RNA sequencing (RNA-seq) analysis from hematological malignancies and normal bone marrow samples. The raw sequencing reads were aligned to human reference genome hg38 using HISAT2, followed by featureCounts quantification, and co-expressed and differentially expressed genes (DEGs) were detected by WGCNA and DESeq2, respectively. Alternative splicing events were calculated by rMATS, and further validated by reverse transcriptase PCR (RT-PCR).
Results
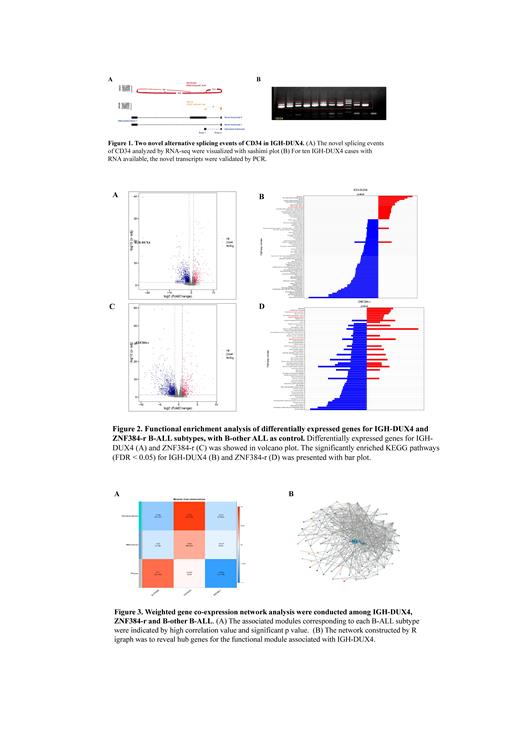
The transcriptome characteristics of hematological malignancies, including AML, B-ALL, T-ALL, and MPAL, were elaborately investigated. We found that two accompanying novel in-frame splicing isoforms of CD34 were exclusively detected in B-ALL. Furthermore, we focused on B-ALL (n = 504) to systematically explore the transcriptional profile of CD34. The two novel splicing isoforms share a common first exon at the 5' untranslated region of CD34 (Figure 1B), suggesting an alternative promoter that mediated the splicing.
We further observe that the novel aberrant CD34 isoforms are mainly accompanied by IGH-DUX4 gene fusion, which has been reported characterized by high expression of CD34 and intragenic ERG deletion. All IGH-DUX4 B-ALL cases (n = 20) in our cohort were positive with the novel aberrant CD34 isoforms and validated by RT-PCR (Figure 1A). We also validated the negative result for the aberrant CD34 isoforms in ZNF384-r (n = 32) B-ALL in our cohort, which is also characterized by high CD34 expression.
According to the RNA-seq based pseudo-time analysis of our B-ALL cohort, IGH-DUX4 cases mostly tended to be common-B ALL, while ZNF384-r cases presented with more Pro-B ALL, which is also in accordance with the previous report based on immunophenotype analysis. To further discover the discrepancy between DUX4-r and ZNF384-r B-ALL subtypes, we compared DEGs and functional enrichment analysis between these two subtypes with B-other ALL as control, respectively (Figure 2). Activation of JAK-STAT signaling and downregulation of SOCS2, which acts as a negative controller of JAK-STAT signaling, were observed in both subtypes (Figure 2). The hematopoietic cell lineage gene set was enriched in ZNF384-r, consistent with its more immature phenotype. The co-expression gene network for IGH-DUX4 was constructed, and the hub gene APELA was emphasized among the closely related genes (Figure 3).
Discussion
In this study, we firstly identified two novel in-frame CD34 isoforms that share a common alternative first exon at its 5'UTR and are highly enriched in the IGH-DUX4 subtype B-ALL. It has been reported that intragenic focal ERG deletion is characteristically co-occurred with IGH-DUX4, and ERG targets CD34 through regulates its super-enhancer to affect the transcription program. It needs further investigation whether ERG deletion contributes to the aberrant transcription of CD34 in IGH-DUX4 subtype B-ALL. We also emphasized APELA as a hub gene in the transcription regulation network of IGH-DUX4. As it currently lacks effective targeted therapy for IGH-DUX4 subtype B-ALL, the potential therapeutic significance of the closely accompanied aberrant CD34 isoforms and APELA as a hub gene deserves further investigation.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal